Beta1 reflects a peptidyl-glutamyl-peptide-hydrolysing-activity (caspase-like, cleavage after acidic amino acids, also termed as “post-glutamyl-peptide hydrolytic” activity), a trypsin-like (after basic amino acids) activity is located at beta2 and a chymotrypsin-like activity (after neutral amino acids) at the beta5-subunit. The ratios of the individual activities are beta5 >> beta2 > beta1. Consequently, in most cases, proteasomal activity is determined via quantification of the beta5 subunits and most proteasomal inhibitors are also aiming for that subunit. Mouse mutants with both beta5 and beta2 knocked out are lethal, while beta5/beta1- and beta1/beta2-knockout-combinations are viable.
The 20S proteasome is able to recognize and degrade oxidatively damaged and (partially) unfolded proteins in an ATP-independent manner.
The exact mechanism of substrate recognition is still unknown, but it was suggested that the alpha rings are able to recognize and bind hydrophobic structures that are usually included in the inside of globular and water-soluble proteins, but can be exposed after (oxidative) damage and (partial) unfolding – a substrate recognition that is similar to that of some shock proteins [
link].
Binding of a substrate induces “gate opening” of the alpha rings, mounting of the substrate into the inside of the proteasome and its proteolytic degradation into short oligopeptides. Those oligopeptides show different lengths in a range from 2-35 amino acids with three maxima: 2-3, 8-10, and 20-30 amino acids.
From an evolutionary point of view, the proteasome is a very old “invention”, found in almost all bacteria (even in archae, the oldest known bacteria at all), plants, mammals, and fungi. The archaeal proteasome shows the same structure like the “modern” one found in mammals like us – 4 stacked rings, each one containing 7 subunits. But in contrast, the alpha-subunits are identical as well as the beta-subunits, each one carrying a proteolytic center (summarizing 14 centers overall).
During evolution, the archaeal proteasome changed into the modern form found in us mammals. Also, several different proteasomal regulators exist, that can bind like attachments to the alpha rings, providing new/extended functions, usually paired with enhanced proteolytic activity.
The most important proteasomal regulator is the so-called 19S. The 20S “core” proteasome can bind one or two 19S regulators, forming the 26S (19S-20S) or the 30S proteasome (19S-20S-19S), respectively. Though, in the literature, both 26S and 30S are mostly summarized under the term “26S proteasome”.
The 19S regulator contains at least 19 different (currently known) subunits: 6 subunits show an ATPase activity (Rpt1-Rpt6), the others do not (Rpn1-12 and Rpn15). The Rpt-subunits form a hexameric ring, that can bind to the alpha-ring of a 20S proteasome, the Rpn-subunits form a lid-like structure, that enables recognition and binding (Rpn10 and -13) of substrates. The whole 19S-complex has a mass of about 1 megadalton, and is larger than the 20S proteasome it binds to.
Please see the following animation (Fig. 05) for more detailed information about the structure of a 26S (here 10)-20S) proteasome.
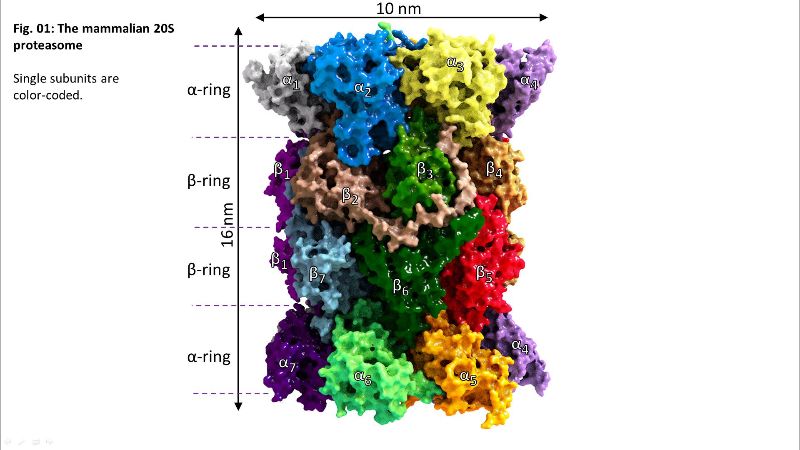 Fig. 01: The 20S proteasome
Fig. 01: The 20S proteasome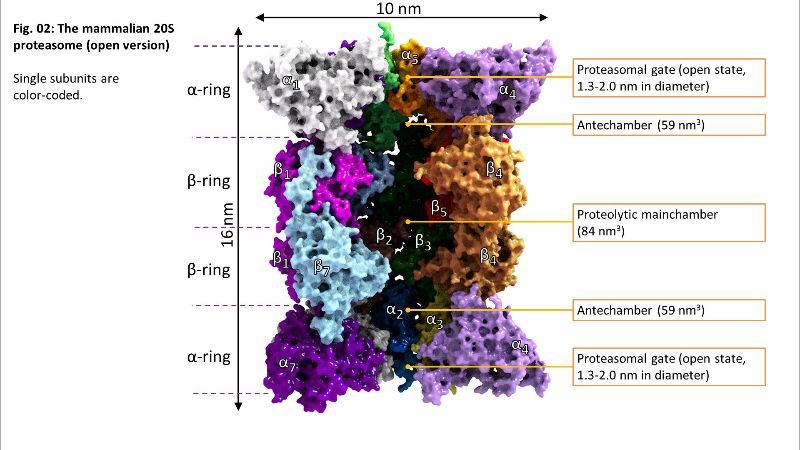 Fig. 02: Inside of 20S
Fig. 02: Inside of 20S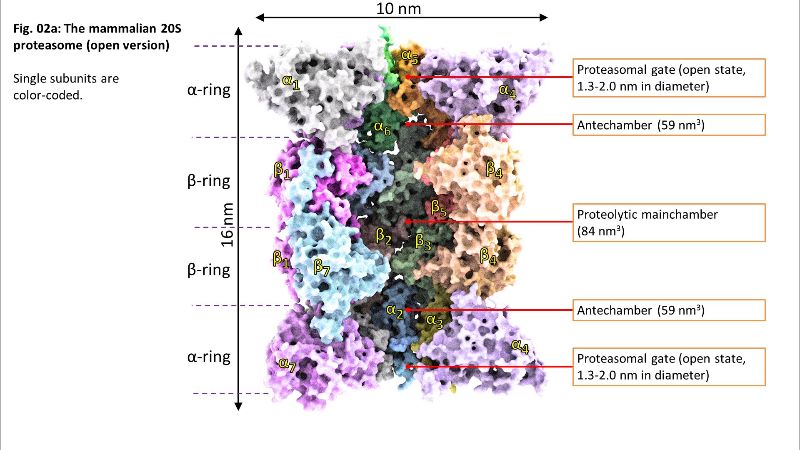 Fig. 02a: Inside of 20S
Fig. 02a: Inside of 20S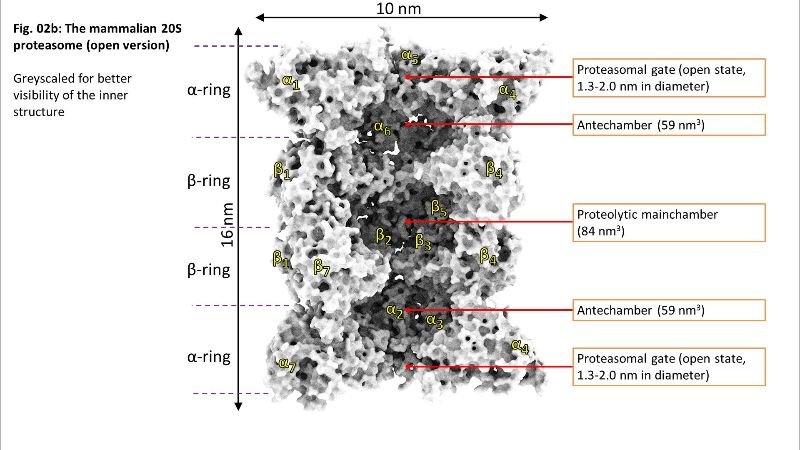 Fig. 02b: Inside of 20S
Fig. 02b: Inside of 20S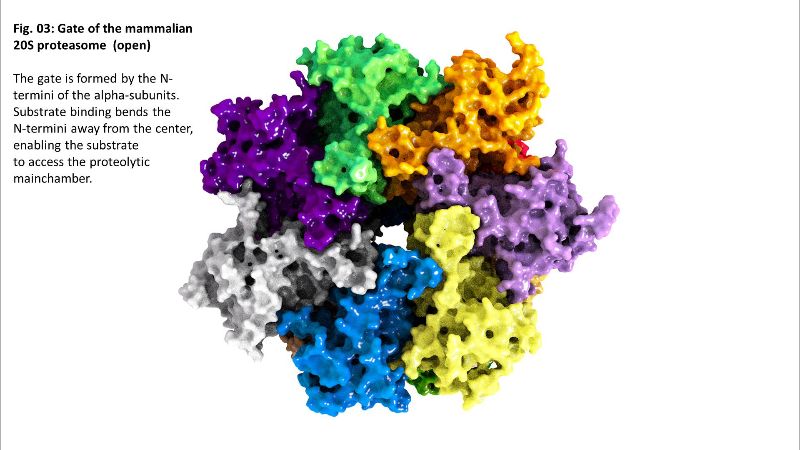 Fig. 03: Gate of 20S
Fig. 03: Gate of 20S